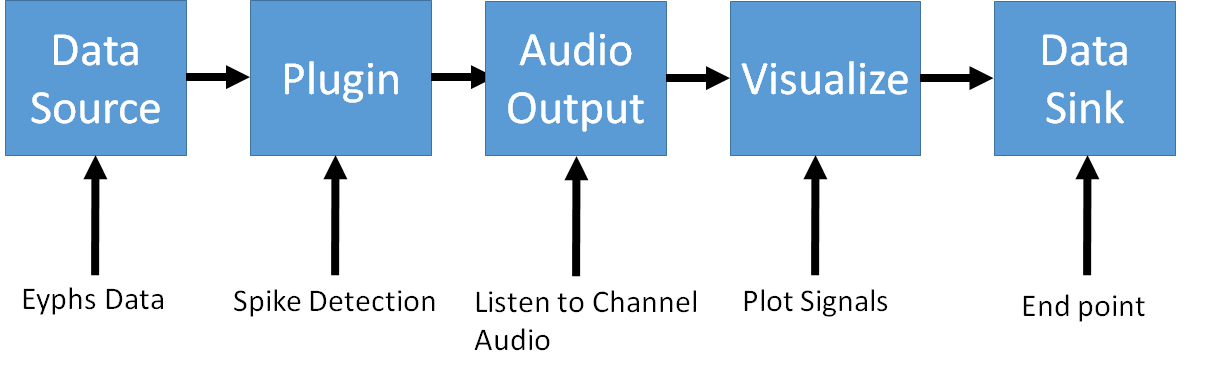
The basic concept of building a data processing pipeline is to connect ready-to-use building blocks (cf: Fig3) to perform several operations one after the other on some given data. Fig2 shows a common series of operations to be performed on electrophysiological data. Every block has an input and at least one output. Data that has been processed by one block is propagated to the output of the next block within the pipeline.
The exception is the Datasink which marks the endpoint of the pipeline and therefore, does not propagate data any further. Its only purpose is to remove data from the computer's memory.
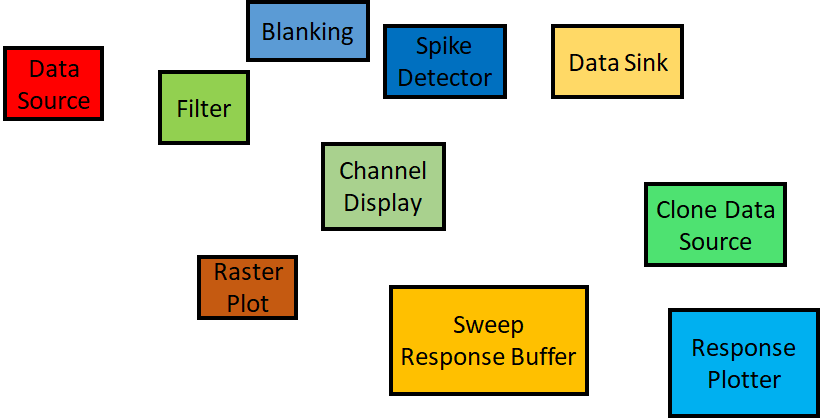
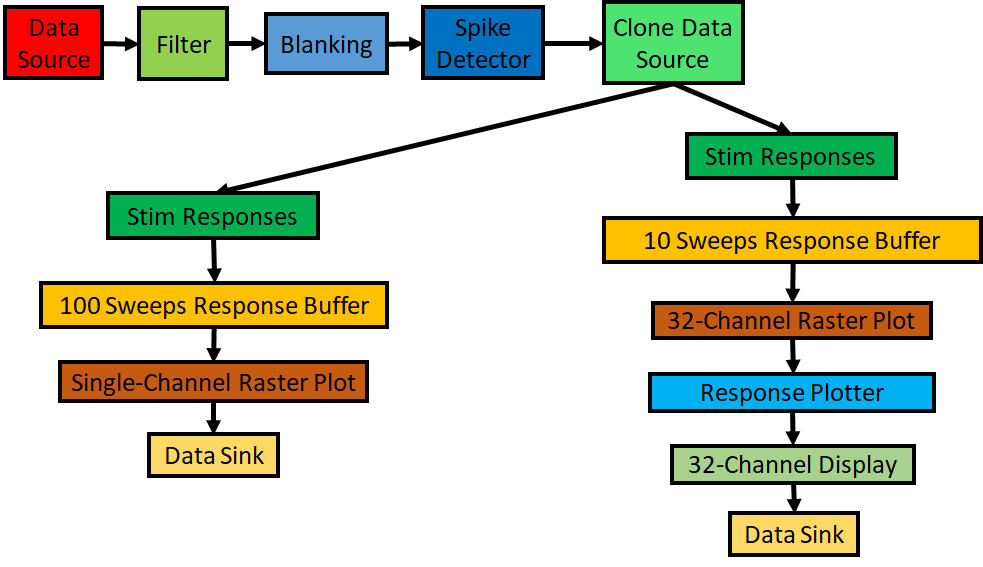
By connecting basic building blocks we can quickly build pipelines that can perform very complex workflows. Over the course of the tutorial, we are going to use all these blocks to build an application for processing electrophysiological data. The pipeline we are going to build is shown in cf: Fig4.
DataFrame
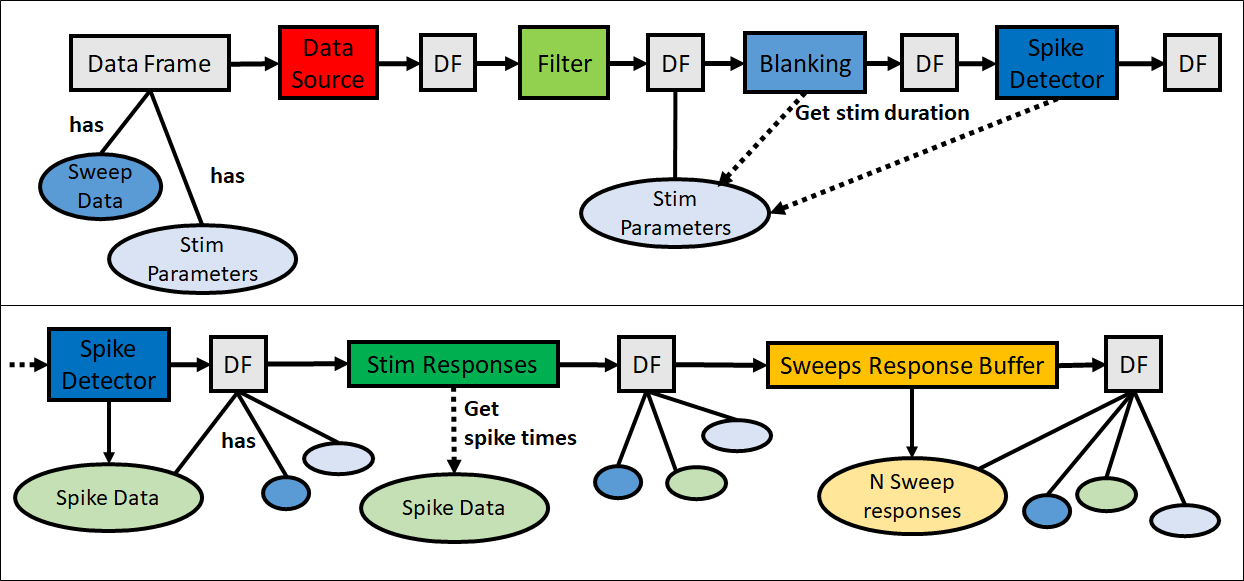
Apart from the Datasource marking the start, and the Datasink marking the endpoint of a pipeline, another special pipeline object is the Dataframe (cf: Fig5). The purpose of the Dataframe is to carry electrophysiological as well as results and meta information E.g. stimulation parameters. The Dataframe is what is passed along the pipeline and what building blocks use to access and store data, results, and other information.